How polarised epithelial cells grow “up” without breaking the barrier
September 28, 2022
Read more
Post-transcriptional modifications are known to influence the fundamental properties and functions of RNAs, with impacts on development, health and disease.
The Kouzarides lab has developed an analysis system based on direct RNA sequencing data from Nanopore, using an approach that compares experimental with unmodified RNA samples, to precisely identify modification sites. They validated the system in vitro and applied it to profile m6A modifications in vivo in yeast and human RNAs.
Leger A et al. (2021) RNA modifications detection by comparative Nanopore direct RNA sequencing. Nat Commun 12, 7198. DOI:10.1038/s41467-021-27393-3.
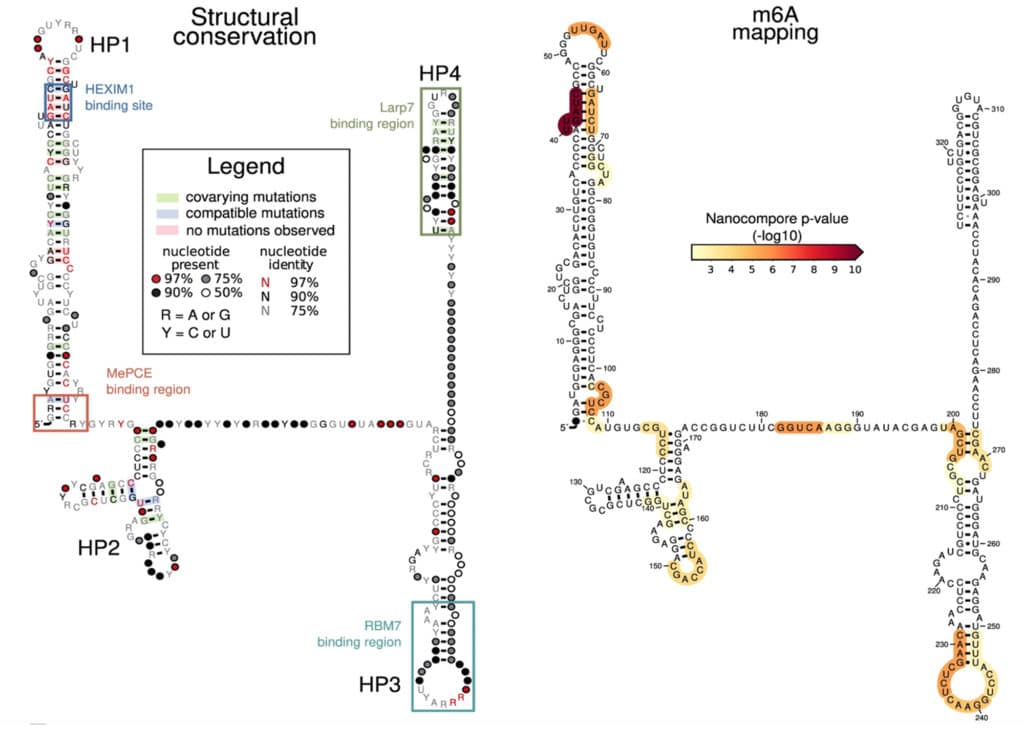
Fig 6A from Leger et al. (2021): m6A identification in 7SK RNA
RNA molecules undergo a vast array of chemical post-transcriptional modifications (PTMs) that can affect their structure and interaction properties. In recent years, a growing number of PTMs have been successfully mapped to the transcriptome using experimental approaches relying on high-throughput sequencing. Oxford Nanopore direct-RNA sequencing has been shown to be sensitive to RNA modifications.
We developed and validated Nanocompore, a robust analytical framework that identifies modifications from these data. Our strategy compares an RNA sample of interest against a non-modified control sample, not requiring a training set and allowing the use of replicates.
We show that Nanocompore can detect different RNA modifications with position accuracy in vitro, and we apply it to profile m6A in vivo in yeast and human RNAs, as well as in targeted non-coding RNAs. We confirm our results with orthogonal methods and provide novel insights on the co-occurrence of multiple modified residues on individual RNA molecules.