New tool to study genome topology and regulatory interactions
December 21, 2021
Read more
Using DNA competition experiments in Xenopus laevis oocytes, Javed et al. show that the stability of gene expression in a non-dividing cell could be caused by the local entrapment of part of the general transcription machinery in transcriptionally active regions. Active and silent forms of the same DNA template are physically separated in the oocyte nucleus in liquid-liquid phase-separated condensates.
Javed K et al. (2022) DNA-induced spatial entrapment of general transcription machinery can stabilise gene expression in a nondividing cell. Proc Natl Acad Sci U S A 119: e2116091119. DOI: 10.1073/pnas.2116091119.
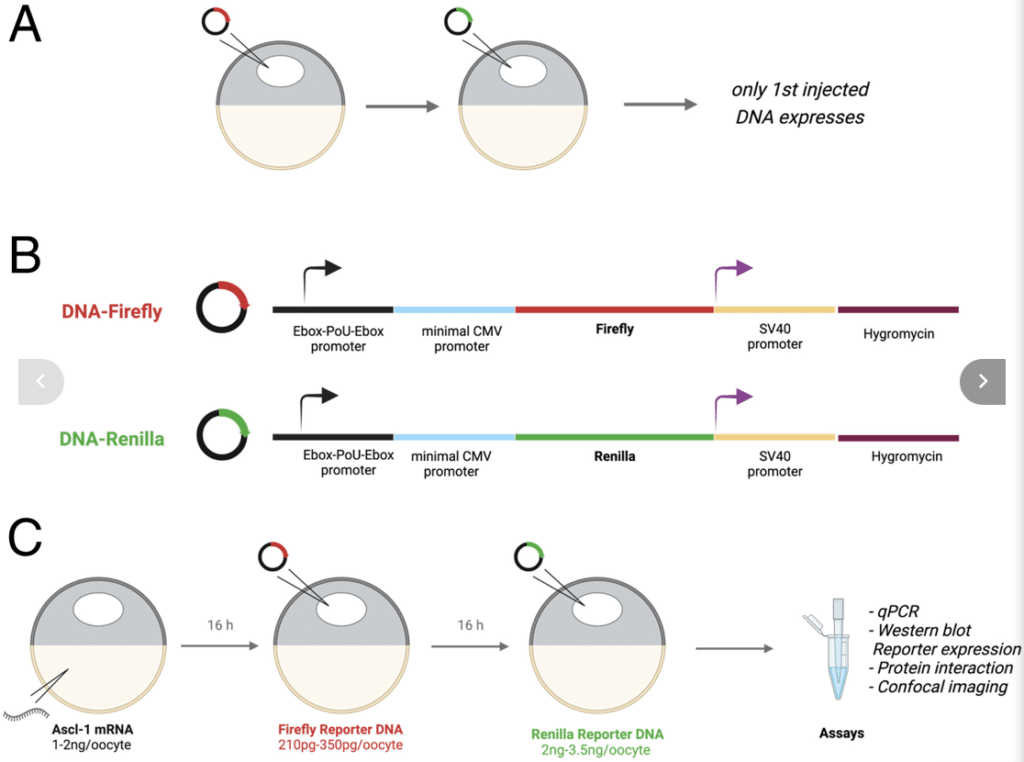
Fig. 1 from the paper shows overall design of the oocyte DNA competition assay.
How differentiated cells such as muscle or nerve maintain their gene expression for prolonged times is currently elusive. Here, using Xenopus oocytes, we have shown that the stability of gene expression in nondividing cells may arise due to the local entrapment of transcriptional machinery to specific gene transcription start sites.
We found that within the same nucleus active versus inactive versions of the same gene are spatially segregated through liquid–liquid phase separation. We further observe that silent genes are associated with RNA-Pol-II phosphorylated on Ser5 but fails to attract RNA-Pol-II elongation factors.
We propose that liquid–liquid phase separation mediated entrapment of limiting transcriptional machinery factors maintain stable expression of some genes in nondividing cells.