Andrea Brand
Head of Wellcome laboratoriesResearch summary
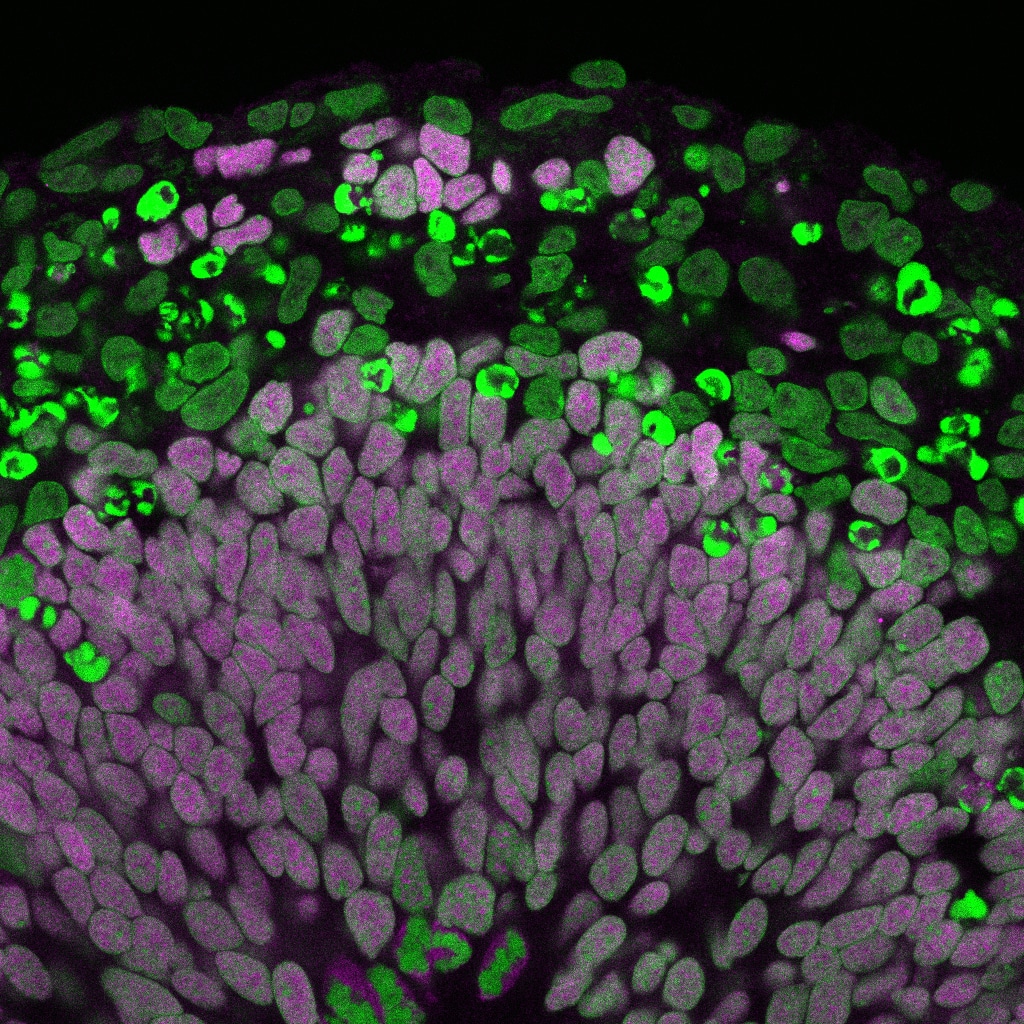
Time to wake up: regulation of stem cell quiescence and proliferation
Stem cell populations in tissues as varied as blood, gut and brain spend much of their time in a mitotically dormant, quiescent, state. A key point of regulation is the decision between quiescence and proliferation. The ability to reactivate neural stem cells in situ raises the prospect of potential future therapies for brain repair after damage or neurodegenerative disease. Understanding the molecular basis for stem cell reactivation is an essential first step in this quest.

Selected publications
-
Agrawal N et al. (2021) Predicting novel candidate human obesity genes and their site of action by systematic functional screening in Drosophila. PLoS Biol 19(11):e3001255. DOI: 10.1371/journal.pbio.3001255.
-
Díaz-Torres A et al (2021) Stem cell niche organization in the Drosophila ovary requires the ECM component Perlecan. Curr Biol 31(8):1744-1753.e5. DOI: 10.1016/j.cub.2021.01.071.
-
Hakes AE & Brand AH (2020) Tailless/TLX reverts intermediate neural progenitors to stem cells driving tumourigenesis via repression of asense/ ASCL1. Elife 9:e53377. DOI: 10.7554/eLife.53377.
-
Otsuki L & Brand AH (2019) Dorsal-ventral differences in neural stem cell quiescence are induced by p57KIP2/Dacapo. Dev Cell 49(2): 293-300.e3. DOI: 10.1016/j.devcel.2019.02.015.
-
Otsuki L & Brand AH (2018) Cell cycle heterogeneity directs the timing of neural stem cell activation from quiescence. Science 360: 99–102. DOI: 10.1126/science.aan8795.
-
Cheetham SW & Brand AH (2018) RNA-DamID reveals cell-type-specific binding of roX RNAs at chromatin-entry sites. Nat Struct Mol Biol 25:109–114. DOI: 10.1038/s41594-017-0006-4.
Biography
Prof Andrea Brand PhD FRS FMedSci
Head of Institute’s Wellcome laboratories, Wellcome Senior Investigator, Herchel Smith Professor of Molecular Biology, Royal Society Darwin Trust Research Professor, Member of the University Department of Physiology, Development and Neuroscience
Research group
-
Catherine Davidson
Research Associate
-
Bernardo Delarue Bizzini
PhD Student
-
Cian Doherty
PhD Student
-
Dr Alex Donovan
Research Associate
-
Laura Gherghina
PhD Student
-
Anna Malkowska
PhD Student
-
Jocelyn Tang
Research Assistant
-
Nemira Zilinskaite
PhD Student