Tony Kouzarides
Senior group leaderResearch summary
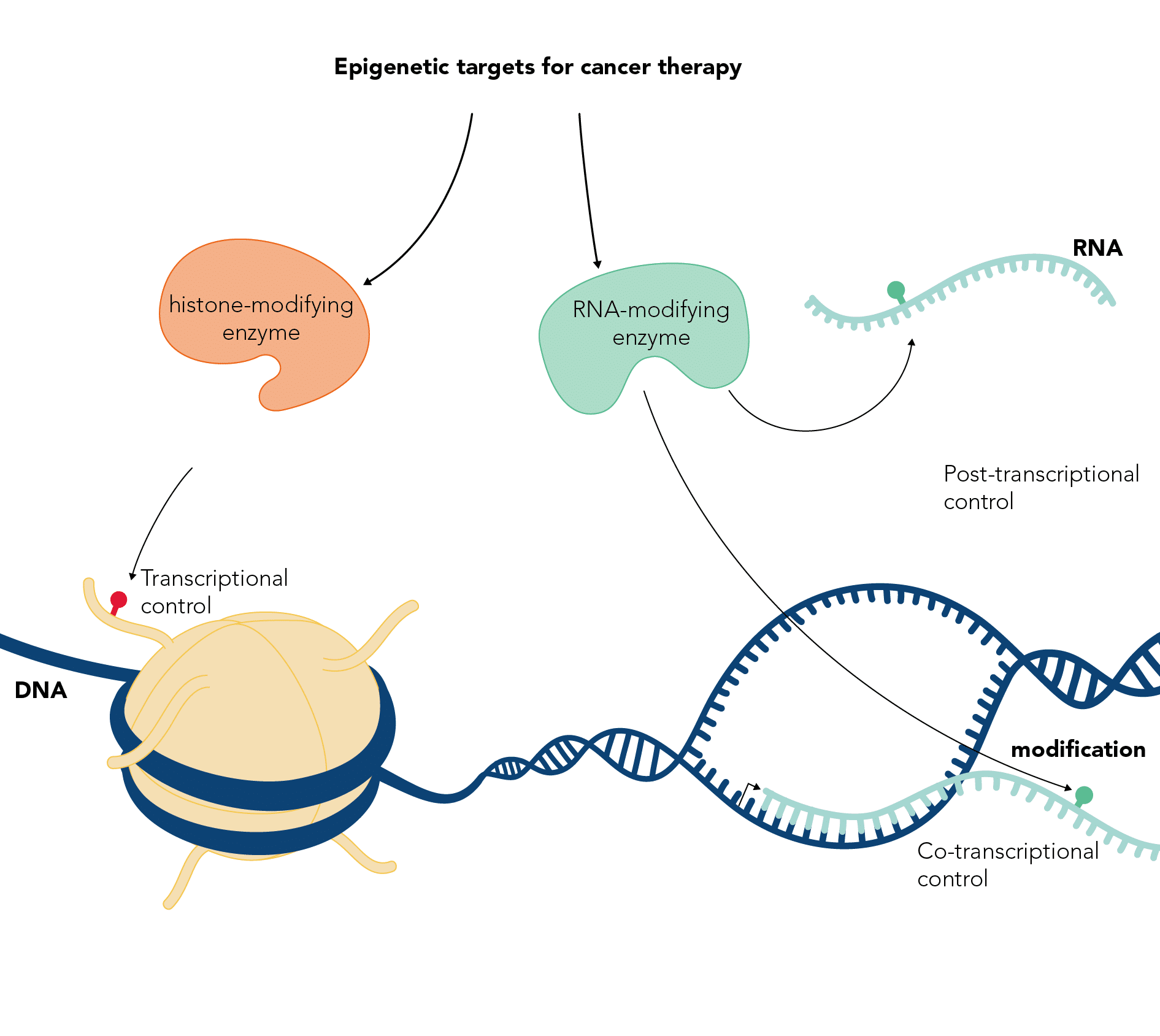
Epigenetic modifications and cancer
DNA exists in the cell nucleus wrapped around histone proteins to form chromatin. The DNA and histones are decorated with many types of covalent chemical modifications, which can affect transcription and other cellular processes. In addition, non-coding RNAs that regulate chromatin function can be similarly chemically modified. Our lab is involved in characterising the pathways that mediate and control DNA, RNA and histone modifications. We try to understand the cellular processes they regulate, their mechanism of action and their involvement in cancer.

Selected publications
-
Leger A et al. (2021) RNA modifications detection by comparative Nanopore direct RNA sequencing. Nat Commun 12: 7198. DOI:10.1038/s41467-021-27393-3.
-
Han N et al. (2021) Identification of SARS-CoV-2-induced pathways reveals drug repurposing strategies. Sci Adv 7(27):eabh3032. DOI 10.1126/sciadv.abh3032.
-
Santos-Rosa H et al. (2021) Methylation of histone H3 at lysine 37 by Set1 and Set2 prevents spurious DNA replication. Mol Cell 81(13):2793-2807.e8. DOI: 10.1016/j.molcel.2021.04.021.
-
Yankova E et al. (2021) Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature 593(7860):597-601. DOI: 10.1038/s41586-021-03536-w.
-
Barbieri I & Kouzarides T (2020) Role of RNA modifications in cancer. Nat Rev Cancer 20, 303–322. DOI: 10.1038/s41568-020-0253-2.
-
Pandolfini L et al. (2019) METTL1 Promotes let-7 MicroRNA Processing via m7G Methylation. Mol Cell Bio 74(6): 1278-1290.e9. DOI: 10.1016/j.molcel.2019.03.040.
-
Barbieri I et al. (2017) Promoter-bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control. Nature 552: 126–131. DOI: 10.1038/nature24678.
Biography
Prof Tony Kouzarides PhD FRS FMedSci
Professor of Cancer Biology, Cancer Research UK Gibb Fellow, Director of the Milner Therapeutics Institute, Member of the University Department of Pathology
Tony did his bachelors’ degree at Leeds University and his PhD at the University of Cambridge. He then did postdoctoral work at the MRC Laboratory of Molecular Biology in Cambridge and at New York University Medical Centre. He returned to the UK to set up his own lab at the Gurdon Institute.
The Kouzarides lab has been studying epigenetic modifications for many years, starting with the identification of the first human enzymes to modify chromatin in 1996. His lab has identified and characterised many chromatin modification pathways and showed their involvement in cancer. The lab is now investigating the functions of mRNA modifications and their connections to cancer. In close collaboration with STORM Therapeutics, the Kouzarides lab is targeting RNA modification pathways with small molecule inhibitors, to develop drugs against cancer.
In the 22-year period of 1996-2017, Tony was listed in the top ten most-cited scientists at the University of Cambridge (in any field), as shown in the curated publication database of 100,000 top scientists world-wide (Ioannidis J et al, Plos Biology, 2019).
Tony is a co-founder and director of the Milner Therapeutics Institute, whose mission is to deliver better therapies through collaborations with industry. He is founder and director of Cambridge Gravity, an organisation for the promotion of science at the University of Cambridge. He is founder and patron of a cancer charity in Spain called Conquer Cancer (Vencer el Cancer). He is on the Scientific Advisory Board of the Institute of Cancer Research (UK) and on the Executive Board of the Cambridge Cancer Centre.
Tony is a co-founder and ex-director of Abcam plc, a publicly trading research reagents company in Cambridge, a co-founder and ex-director of Chroma Therapeutics, a drug discovery company based in Oxford, and a co-founder and current director of STORM Therapeutics, a drug discovery company based in Cambridge.
Notable achievements and honours
-
2021Nemitsas Prize
-
2020Cyprus Excellence in Science Award
-
2020Member of the Cyprus Academy of Sciences, Letters and Arts
-
2020Member of the American Academy of Arts and Sciences
-
2013Biochemistry Society Novartis Medal and Prize
-
2013Heinrich Wieland Prize
-
2012Fellow of the Royal Society
-
2003Wellcome Trust Award for Research Related to Medicine
-
2001Member of the Academy of Medical Sciences
-
1996Bodossaki Foundation Prize
Research group
-
Dr Andrew Bannister
Senior Research Associate
-
Dr Alex Casanova
Research Associate
-
Lauren Harrison-Oakes
Part III Student
-
Marie Klimontova
PhD Student
-
Sri Lestari
Senior Research Laboratory Technician
-
Alexandra Michaelidou
PhD Student
-
Dr Carlos Melo
Research Associate
-
Dr Helena Santos Rosa
Senior Research Associate
-
Han Zhang
Bioinformatician