Mitoballs, large mitochondrial clusters, form to sustain sperm development
August 14, 2023
Read more
Development of the mammalian germline entails epigenetic resetting, including the loss of DNA methylation, which is essential for germline maturation, totipotency and embryonic development in the next life cycle. Gruhn et al. find that the loss of repressive epigenetic information in the early human germline extends to major repressive histone modifications. Consequently, efficient maintenance of a heterochromatic state is limited to a subset of loci, including some developmental genes and evolutionarily young transposable elements. Notably, reducing the repressive chromatin state in a substantial portion of the genome could promote a purifying selection against hPGCs harbouring aberrantly active transposable elements.
Gruhn W. et al. (2023) Epigenetic resetting in the human germ line entails histone modification remodeling. Science Advances 2023 Jan 18;9(3):eade1257. DOI: 10.1126/sciadv.ade1257.
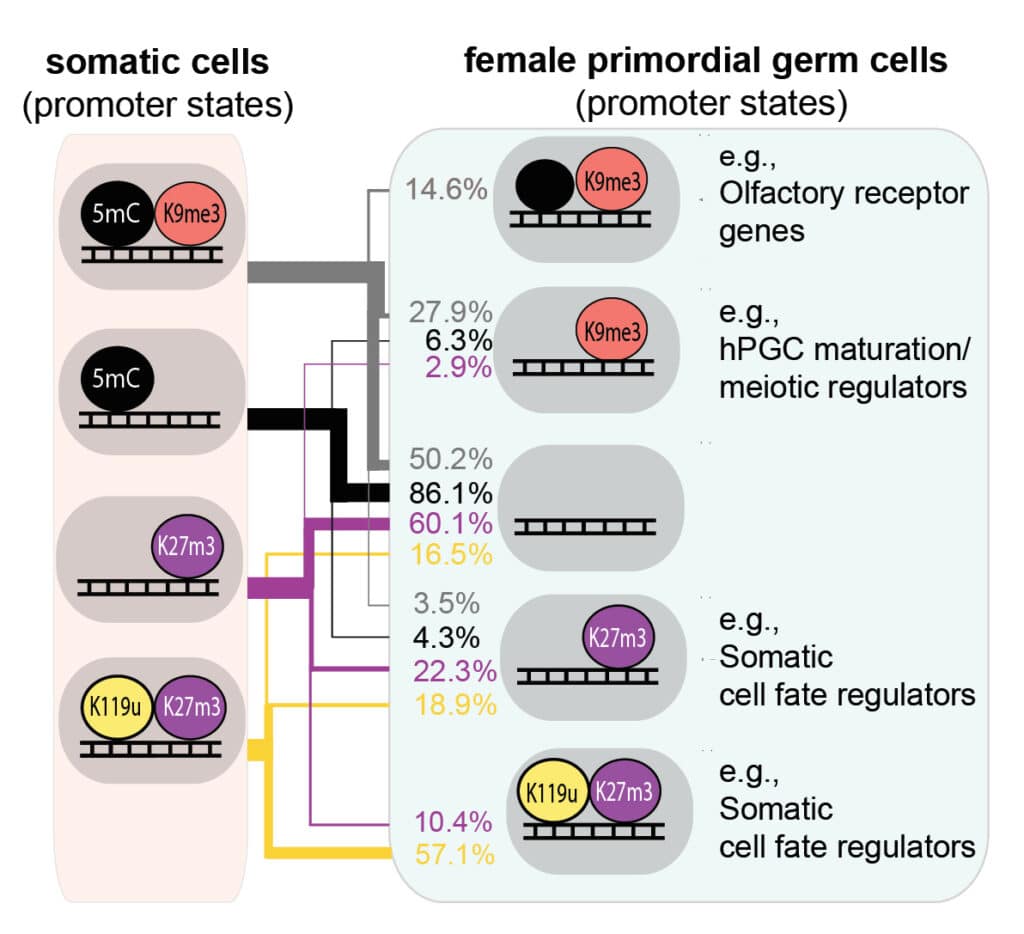
Epigenetic resetting in the mammalian germline entails acute DNA demethylation, which lays the foundation for gametogenesis, totipotency and embryonic development. Here, we characterize the epigenome of hypomethylated human primordial germ cells (hPGCs) to reveal mechanisms preventing the widespread de-repression of genes and transposable elements (TEs). Along with the loss of DNA methylation, we show that hPGCs exhibit a profound reduction of repressive histone modifications resulting in diminished heterochromatic signatures at most genes and TEs and the acquisition of a neutral or paused epigenetic state without transcriptional activation. Efficient maintenance of a heterochromatic state is limited to a subset of genomic loci, such as evolutionarily young TEs and some developmental genes, which require H3K9me3 and H3K27me3, respectively, for efficient transcriptional repression. Accordingly, transcriptional repression in hPGCs presents an exemplary balanced system relying on local maintenance of heterochromatic features and a lack of inductive cues.