Detecting RNA modifications
December 10, 2021
Read more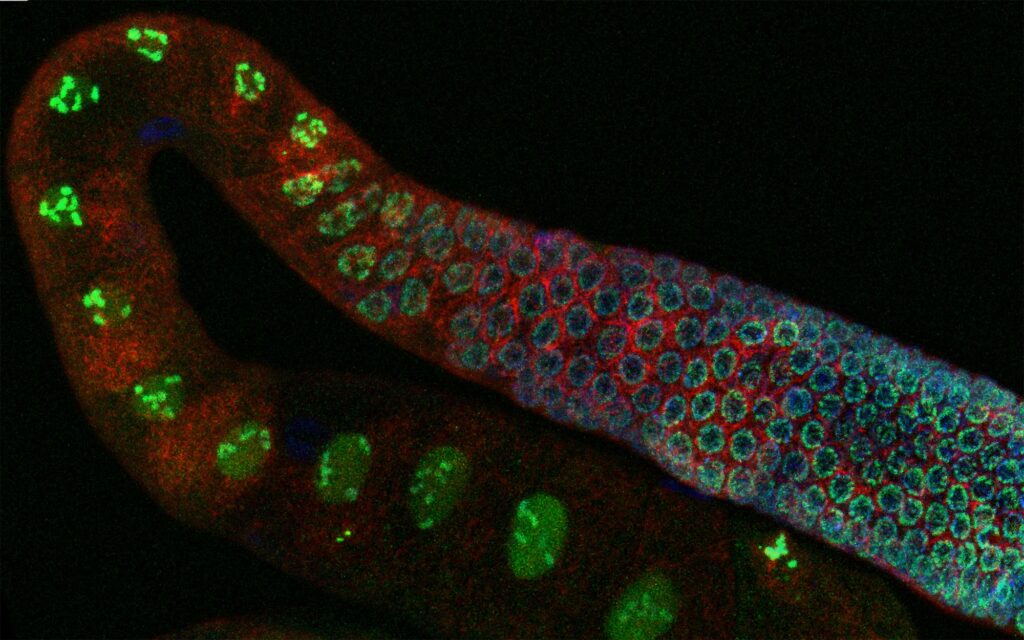
Carelli et al. find that two families of transposable elements (TEs) have been co-opted as germline-specific promoters in Caenorhabditis elegans. HIM-17, a transcription factor evolutionarily related to TE-encoded transposases regulates the expression of co-opted promoters. By tracing the evolutionary history of the co-option events, they found that most are shared with the distantly-related C. briggsae, though species-specific promoters are also found in both nematodes. This work provides solid evidence of large-scale transcriptional network re-wiring through the co-option of TE-derived promoters and their evolutionary preservation.
Carelli et al. (2022) Widespread transposon co-option in the Caenorhabditis germline regulatory network. Science Advances 8(50):eabo4082. DOI: 10.1126/sciadv.abo4082
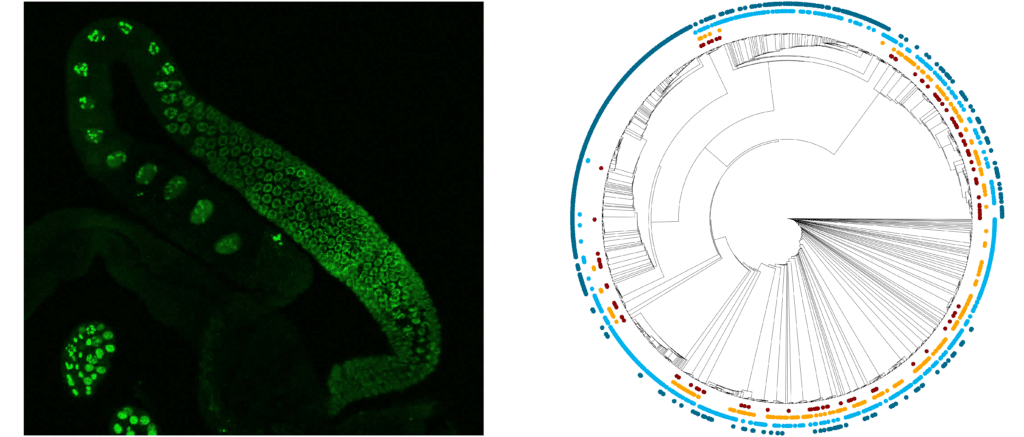
Left: GFP expression in C. elegans germline driven by a CERP2-derived promoter; right: evolutionary relationships between regulatory active (red and yellow) or inactive (light/dark blue) CERP2 repeats.
The movement of selfish DNA elements can lead to widespread genomic alterations with potential to create novel functions. We show that transposon expansions in Caenorhabditis nematodes led to extensive rewiring of germline transcriptional regulation. We find that about one-third of Caenorhabditis elegans germline-specific promoters have been co-opted from two related miniature inverted repeat transposable elements (TEs), CERP2 and CELE2. These promoters are regulated by HIM-17, a THAP domain–containing transcription factor related to a transposase. Expansion of CERP2 occurred before radiation of the Caenorhabditis genus, as did fixation of mutations in HIM-17 through positive selection, whereas CELE2 expanded only in C. elegans. Through comparative analyses in Caenorhabditis briggsae, we find not only evolutionary conservation of most CERP2 co-opted promoters but also a substantial fraction that are species-specific. Our work reveals the emergence and evolutionary conservation of a novel transcriptional network driven by TE co-option with a major impact on regulatory evolution.