Tracing the origin and segregation of the human germline
May 22, 2023
Read more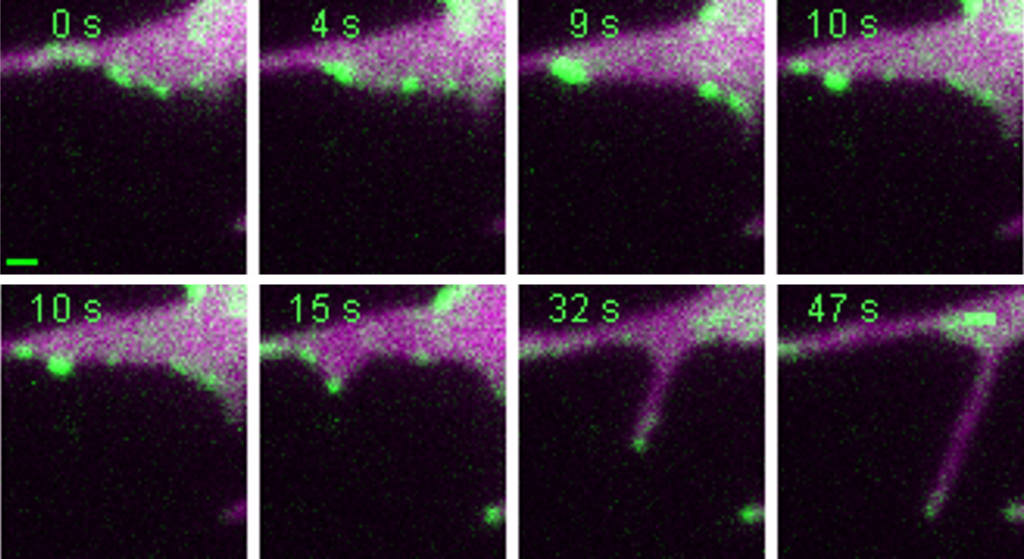
In their review the researchers explore recent advances in conceptual models of how filopodia form, the molecules involved in this process, and their latest understanding of filopodia from in vitro reconstitution and in vivo imaging.
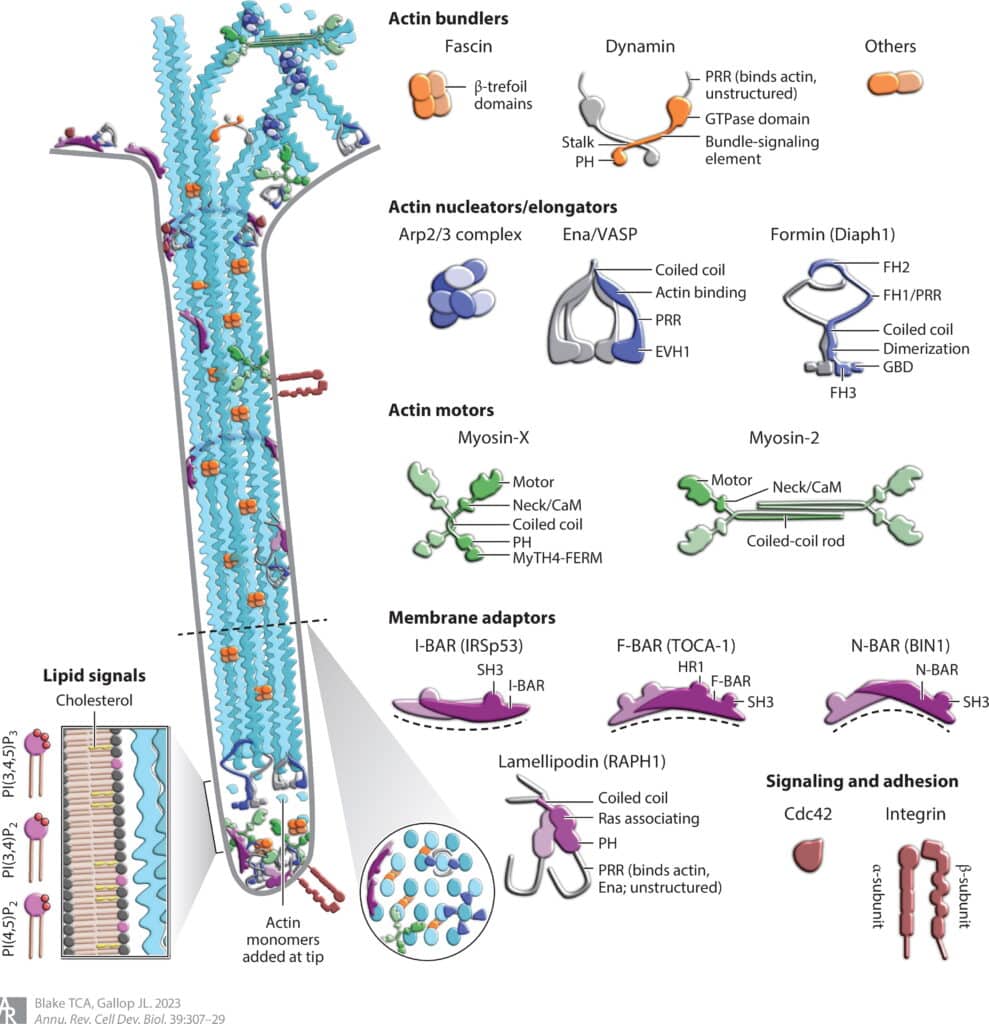
Protein and lipid organization in filopodia
Filopodia are dynamic cell surface protrusions used for cell motility, pathogen infection, and tissue development. The molecular mechanisms determining how and where filopodia grow and retract need to integrate mechanical forces and membrane curvature with extracellular signaling and the broader state of the cytoskeleton. The involved actin regulatory machinery nucleates, elongates, and bundles actin filaments separately from the underlying actin cortex. The refined membrane and actin geometry of filopodia, importance of tissue context, high spatiotemporal resolution required, and high degree of redundancy all limit current models. New technologies are improving opportunities for functional insight, with reconstitution of filopodia in vitro from purified components, endogenous genetic modification, inducible perturbation systems, and the study of filopodia in multicellular environments. In this review, we explore recent advances in conceptual models of how filopodia form, the molecules involved in this process, and our latest understanding of filopodia in vitro and in vivo.