NanoDam finds missing temporal factors
May 9, 2022
Read more
The St Johnston lab’s single-molecule localisation microscopy method for imaging tissues has revealed that nuclear pores are not evenly distributed in the nuclear envelopes of most Drosophila tissues, as they are in tissue culture cells.
Cheng J et al. (2021) A single-molecule localization microscopy method for tissues reveals nonrandom nuclear pore distribution in Drosophila. J Cell Sci 134 (24): jcs259570. DOI: 10.1242/jcs.259570.
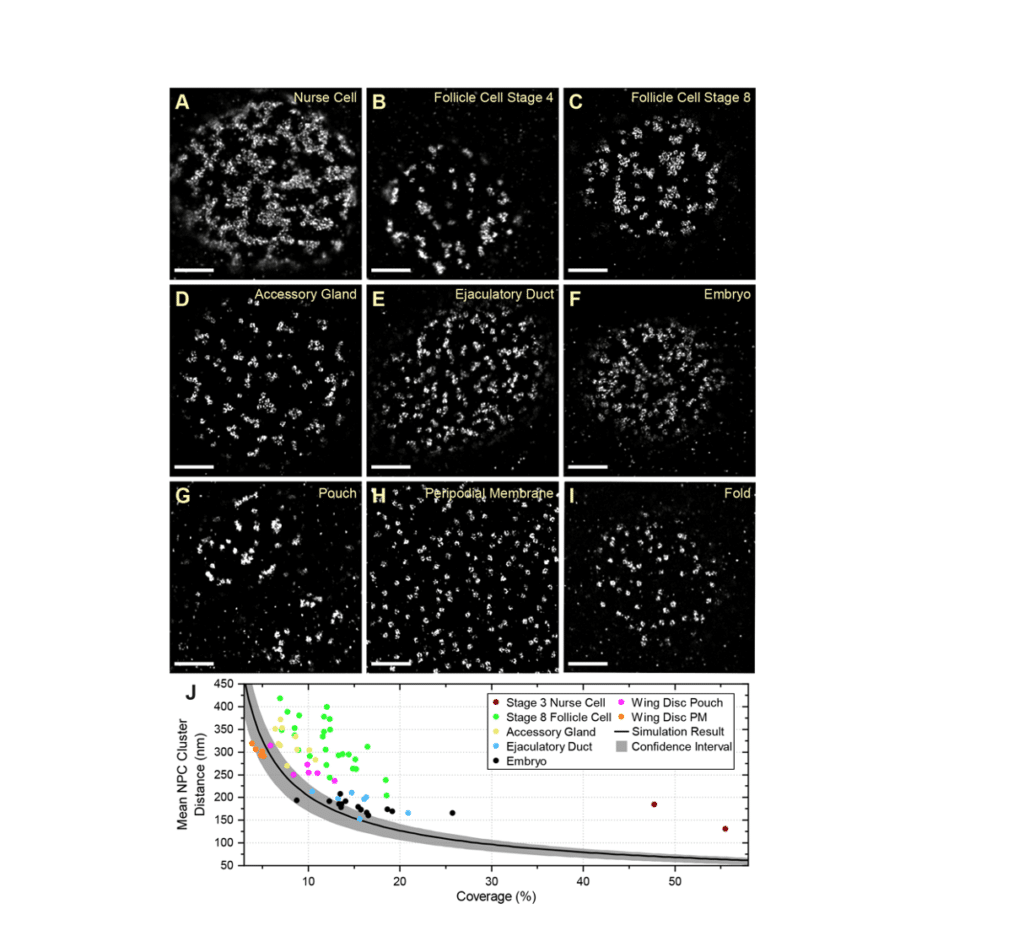
Fig. 6 from Cheng et al (2021)
Single-molecule localization microscopy (SMLM) can provide nanoscale resolution in thin samples but has rarely been applied to tissues because of high background from out-of-focus emitters and optical aberrations.
Here, we describe a line scanning microscope that provides optical sectioning for SMLM in tissues. Imaging endogenously-tagged nucleoporins and F-actin on this system using DNA- and peptide-point accumulation for imaging in nanoscale topography (PAINT) routinely gives 30 nm resolution or better at depths greater than 20 µm. This revealed that the nuclear pores are nonrandomly distributed in most Drosophila tissues, in contrast to what is seen in cultured cells.
Lamin Dm0 shows a complementary localization to the nuclear pores, suggesting that it corrals the pores. Furthermore, ectopic expression of the tissue-specific Lamin C causes the nuclear pores to distribute more randomly, whereas lamin C mutants enhance nuclear pore clustering, particularly in muscle nuclei. Given that nucleoporins interact with specific chromatin domains, nuclear pore clustering could regulate local chromatin organization and contribute to the disease phenotypes caused by human lamin A/C laminopathies.