Unlocking the potential for clinical application of reprogrammed somatic cells by germinal vesicles from Xenopus meiotic oocytes
March 1, 2024
Read more
The Ahringer lab present Accessible Region Conformation Capture (ARC-C), a method they have developed that profiles interactions between regulatory elements at high resolution in a single genome-wide assay. Applied to C. elegans, ARC-C revealed unappreciated domain and compartment structure and proteins that are candidates for mediating organisation.
Huang N et al. (2021) Accessible Region Conformation Capture (ARC-C) gives high resolution insights into genome architecture and regulation. Genome Res gr.275669.121. DOI: 10.1101/gr.275669.121. Online in advance.
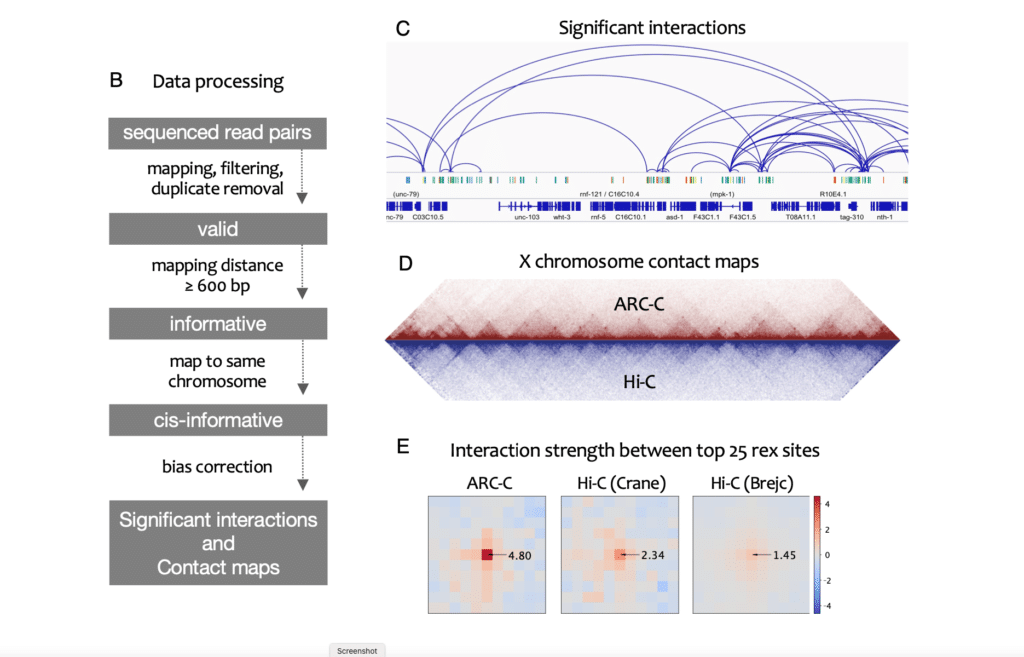
Diagram of data processing steps in ARC-C method of Huang et al. 2021 (Fig 1B)
Nuclear organization and chromatin interactions are important for genome function, yet determining chromatin connections at high-resolution remains a major challenge. To address this, we developed Accessible Region Conformation Capture (ARC-C), which profiles interactions between regulatory elements genome-wide without a capture step.
Applied to C. elegans, we identify ~15,000 significant interactions between regulatory elements at 500bp resolution. Of 105 TFs or chromatin regulators tested, we find that the binding sites of 60 are enriched for interacting with each other, making them candidates for mediating interactions. These include cohesin and condensin II.
Applying ARC-C to a mutant of transcription factor BLMP-1 detected changes in interactions between its targets. ARC-C simultaneously profiles domain level architecture, and we observe that C. elegans chromatin domains defined by either active or repressive modifications form topologically associating domains (TADs) which interact with A/B (active/inactive) compartment-like structure. Furthermore, we discovered that inactive compartment interactions are dependent on H3K9 methylation.
ARC-C is a powerful new tool to interrogate genome architecture and regulatory interactions at high resolution.