Unexpected clustering of nuclear pores revealed by new tissue imaging system
December 16, 2021
Read more
The Brand lab apply Targeted DamID (TaDa) to the developing mouse cortex by in-utero electroporation to find the network of genes that are the targets of NOTCH and its transcription factor RBPJ. The researchers then show that repression of NSC-specific Notch target genes in intermediate progenitors and neurons correlates with decreased chromatin accessibility.
van den Ameele, J et al. (2022) Reduced chromatin accessibility correlates with resistance to Notch activation. Nat Commun 13, 2210. DOI: 10.1038/s41467-022-29834-z.
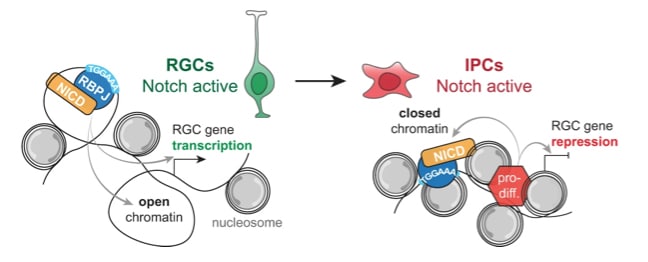
Fig 5h. Model of NOTCH target gene repression during differentiation after ectopic expression of NICD-Dam.
The Notch signalling pathway is a master regulator of cell fate transitions in development and disease. In the brain, Notch promotes neural stem cell (NSC) proliferation, regulates neuronal migration and maturation and can act as an oncogene or tumour suppressor. How NOTCH and its transcription factor RBPJ activate distinct gene regulatory networks in closely related cell types in vivo remains to be determined.
Here we use Targeted DamID (TaDa), requiring only thousands of cells, to identify NOTCH and RBPJ binding in NSCs and their progeny in the mouse embryonic cerebral cortex in vivo. We find that NOTCH and RBPJ associate with a broad network of NSC genes. Repression of NSC-specific Notch target genes in intermediate progenitors and neurons correlates with decreased chromatin accessibility, suggesting that chromatin compaction may contribute to restricting NOTCH-mediated transactivation.