Roadmap to guide in vitro gonadogenesis
July 6, 2022
Read more
George Sirinakis and colleagues in the St Johnston lab explore the performance of four spinning disk geometries for super-resolution imaging. The aim is to optimise for thicker biological samples in which PAINT is being used for single molecule localisation, in this case tracking actin in the Drosophila egg chamber. Disk geometries with higher light collection efficiency performed the best.
Sirinakis G et al. (2022) Quantitative comparison of spinning disk geometries for PAINT based super-resolution microscopy. Biomedical Optics Express 13 (7): 3773-3785. DOI: 10.1364/BOE.459490.
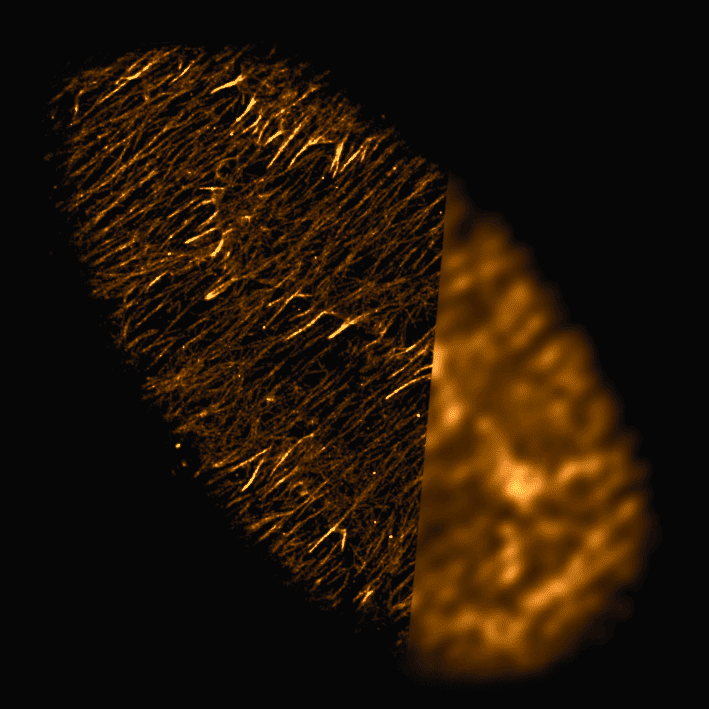
PAINT methods that use DNA- or protein- based exchangeable probes have become popular for super-resolution imaging and have been combined with spinning disk confocal microscopy for imaging thicker samples. However, the widely available spinning disks used for routine biological imaging are not optimized for PAINT-based applications and may compromise resolution and imaging speed.
Here, we use Drosophila egg chambers in the presence of the actin-binding peptide Lifeact to study the performance of four different spinning disk geometries. We find that disk geometries with higher light collection efficiency perform better for PAINT-based super-resolution imaging due to increased photon numbers and, subsequently, detection of more blinking events.